Abstract
Background: Structural variants (SV) are known to play a critical role in the pathogenesis of multiple cancer types. Using whole genome sequencing (WGS), we recently characterized the SV landscape of 752 newly diagnosed multiple myeloma (MM) patients. We defined 68 SV hotspots involving key driver genes, recurrent copy number variations (CNV), and aberrant gene expression (Rustad et al. Blood Cancer Discovery 2020). Despite this comprehensive annotation, more than half of SVs were not linked to any known recurrent MM genomic driver event and were left undefined. The biological impact of these "rare SV" events, cumulatively occurring in 93% (702/752) of patients, is largely unknown.
Methods: To study the biological impact of rare SVs, we interrogated WGS (n=752) and RNAseq (n=591, 78.6%) in the CoMMpass trial. Recurrent SV were excluded, identified by involvement in any of canonical translocations (i.e. Ig translocations), recurrent CNV identified by GISTIC (n=152), and SV hotspots (n= 68). If both breakpoints of an SV were not involved in a recurrent event, the SV was identified as rare. Complex events such as chromothripsis were defined rare if all SV breakpoints involved were rare.
To infer the biological impact of each rare SV and distinguish their role between a passenger or potential driver, we investigated their impact on gene expression in a direct manner - where an SV occurs within a gene body - and indirect manner - where they function through transcriptional deregulation via superenhancer hijacking. SVs were considered involved with a gene if either breakpoint occurred up to 1 Mb from/or intersects the gene region. To determine SV class-specific breakpoint enrichment in relation to distance from genes, a re-shuffle permutation was performed to create a random background model for each SV class and gene expression direction (up vs down regulated). Extending on methods developed to test the penetrance of rare germline events (Chiang et al. Nat Gen 2017), genes paired to rare SVs were considered affected if the gene expression was above a gene specific outlier Z-score of +/- 2. Lastly, we interrogated the indirect link between rare SVs and gene expression outliers, modeling breakpoint density to the nearest known MM superenhancer, up to 10 MB. To determine if this association was higher than what expected by chance, rare SV breakpoints were re-shuffled, adding a random length between 10-20 MB to the original position, and proximity to nearest superenhancer was re-calculated.
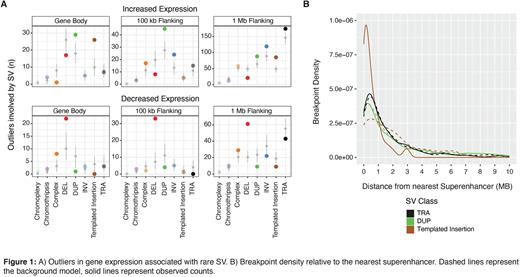
Results: Of the total 8,942 SVs, 726 (8%) were involved with canonical MM translocations, 1,453 (16%) involved in SV hotspots, and 2,014 (22%) involved with GISTIC CNV, leaving 4,749 (53%) rare SVs. Rare templated insertions were enriched in overexpressing outliers within the body of genes and up to 1 MB away. Rare duplication SVs were enriched in overexpressing outliers within the gene body, in genes 100 kb of the breakpoint and up to 1 MB away. Rare inversion SVs were enriched in overexpressing outliers of genes 100kb and 1MB away, and rare translocations were associated with overexpressing outliers 1 MB away (Fig 1a). Rare complex SVs were enriched within the gene body of underexpressing outliers, while deletion SVs were enriched in gene outliers with decreased expression within the gene body, 100 kb away, and up to 1 MB away (Fig 1a). A total of 201 (34%) patients had at least 1 rare SV event associated with gene expression outliers and, interestingly, these patterns of enrichment of rare SVs were comparable to patterns of enrichment observed across recurrent SV events.
Rare duplications, translocations and templated insertions were significantly enriched within and/or up to 1 MB from superenhancer regions, with templated insertion significantly enriched against the background model (p < 0.001, Fisher's Exact test) (Fig 1b). Overall, 82% (104/126) of gene outliers affected by rare templated insertions were associated with superenhancers, 54% (130/237) by rare translocations, and 55% (96/172) by rare duplication events.
Conclusion: Leveraging WGS and RNA-seq of clinical samples, we demonstrate that rare SVs, which collectively account for most SVs observed in MM, are frequently associated with aberrant gene expression and can play a potential driver role in MM pathogenesis. These data further expand our understanding of immense MM heterogeneity and may have significant implications for the development of individualized treatment.
Disclosures
Usmani:Abbvie, Amgen, BMS, Celgene, EdoPharma, Genentech, Gilead, GSK, Janssen,Oncopeptides, Sanofi, Seattle Genetics, SecuraBio, SkylineDX, Takeda, TeneoBio: Consultancy; Amgen, BMS, Janssen, Sanofi: Speakers Bureau; Amgen, Array Biopharma, BMS, Celgene, GSK, Janssen, Merck, Pharmacyclics, Sanofi, Seattle Genetics, SkylineDX, Takeda: Research Funding. Landgren:Theradex: Other: Independent Data Monitoring Committee (IDMC) member for clinical trials; Pfizer Inc: Consultancy; NCI/NIH: Research Funding; Riney Foundation: Research Funding; Tow Foundation: Research Funding; Rising Tide Foundation: Research Funding; Leukemia & Lymphoma Society: Research Funding; MMRF: Honoraria; Aptitude Health: Honoraria; Merck & Co., Inc.: Other: Independent Data Monitoring Committee (IDMC) member for clinical trials; Janssen: Honoraria, Other: Independent Data Monitoring Committee (IDMC) member for clinical trials, Research Funding; Amgen: Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal